Genomic code that makes us is made up of four letters, ATGC. Billions of these letters together creates a lifeform. Iterated function systems (IFS) are anything that can be made by repeating the same simple rules over and over. The easiest example being tree branches, add a simple structure repeatedly ad-infinitum and before you know it we have complex and beautiful systems; the popular example being the Sierpinski Triangle or “triforce” for the Zelda fans. As the cost of DNA sequencing becomes cheaper day by day we are confronted with a tsunami of data and it has become exceedingly difficult to derive meaningful answers from all the information contained within us.
Finding any advantage in ways to organize and view the data helps us discover minuet differences between individuals or say a normal cell versus a cancer cell. This is where Chaos Game Representation (CGR) becomes helpful, CGR is just a form of IFS that is helpful in mapping seemingly random information, that we suspect or know to have some sort of underlying structure.
In our case this would be the human genome. Although when looking at the letters coming from our DNA it seems like billions of random babbles, it is of course organized in a manner to give the blueprint for our bodies. So let’s roll the dice- do we get any sort of meaningful structure when applying CGR to DNA? If you are so inclined, something fun to try is the following:
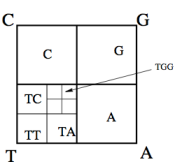
genome = Import["c:\data\sequence.fasta", "Sequence"]; genome = StringReplace[ToString[genome], {"{" -> "", "}" -> ""}]; chars = StringCases[genome, "G" | "C" | "T" | "A"]; f[x_, "A"] := x/2; f[x_, "T"] := x/2 + {1/2, 0}; f[x_, "G"] := x/2 + {1/2, 1/2}; f[x_, "C"] := x/2 + {0, 1/2}; pts = FoldList[f, {0.5, 0.5}, chars]; Graphics[{PointSize[Tiny], Point[pts]}]
For example, reading the sequence in order, apply T1 whenever C is encountered, apply T2 whenever A is encountered, apply T3 whenever T is encountered, and apply T4 whenever G is encountered. Really though any transformations to C, A, T, and G can be used and multiple methods can be compared. Self-similarity is immediately noticeable in these maps, which isn’t all that surprising since fractals are abundant in nature and DNA after all, is a natural syntax. Being aware that these patterns exist within our data, opens us up to some new questions to evaluate if IFS, CGR and fractals in general are helpful tools in the interpretation of genomic data.
Since the mapping is 1-1 and we see patterns emerge, we are hinted that there may be biological relevance; especially because different genes yield different patterns. But what exactly are the correlations between the patterns and the biological functions? It would also be very interesting to see mappings of introns/exons colored differently or color amino acids and various codons. One thing is for sure, genomes aren’t just endless columns and rows of letters, they are pictures. It is much easier to compare pictures and discover variations, which can ultimately allow us to find meaningful interpretation from this invaluable data.
Citations:
Jeffrey, H. J., “Chaos game visualization of sequences,” Computers & Graphics 16 (1992), 25-33.
Ashlock, D. Golden, J.B., III. Iterated function system fractals for the detection and display of DNA reading frame (2000) ISBN: 0-7803-6375-2
VV Nair, K Vijayan, DP Gopinath ANN based Genome Classifier using Frequency Chaos Game Representation (2010)








Awesome post! It inspired this little simple web app
http://codepen.io/alojzije/full/xfmKr
nice!
Pingback: Gene Visualization | Petri Dish Talk
Some years ago (2006), I’ve done my master thesis [2] about chaos game representation of DNA sequences … and found that those fractal patterns are the representation of “regular languages” [1] embeded in the DNA sequence and proposed a way to filter the representation by removing the patterns. Then, you are allowed to mine the genome and search more precice statistical biais.
The CGR have been used to classify genomes between species : use a probabilistic automata [2], compute an euclidian distance or a principal component analysis [3], use a neural network (cf. your links) or whatever classifying method you want.
My feeling is that the CGR can be usefull to produce beautiful pictures and perhaps to illustrate some result. But everything you can do with CGR can be done without CGR (CGR is just a way to represent word frequencies and organize them in a 2d square). For example, suffix arrays are a far more efficient method to compute those frequencies.
If you have any question about my repport [2], ask me 😉
[1] http://en.wikipedia.org/wiki/Regular_language
[2] ftp://ftp.irisa.fr/local/caps/DEPOTS/RapportsStages2006/Rapport_Henin_Thibaut.pdf
[3] http://mbe.oxfordjournals.org/content/16/10/1391.full.pdf
similar triangles.
beautiful
A bioinformatics article which is both readable, informative and concise.
Brilliant. And the insight on images? Agree completely.